
I serves as Director of Bioinformatics and Research Computing at the Duke University School of Medicine and am a professor in the Biostatistics & Bioinformatics Department.
I am not currently accepting students or postdoctoral fellows.
GoogleScholar Full cv and publication list
I am an expert in bioinformatics, cancer genomics, and genomics with a computer science PhD and 20 years’ track record of impactful research as evidenced by a Google Scholar h-index of 85 and i10-index of 165. I have extensive experience in leading wet-lab as well as computational research.
I founded the Duke-NUS Centre for Computational Biology, whose 5 faculty (including me) published > 250 papers between 2016 and 2023 and held US$ 10 million in competitive funding in 2023. In addition to the faculty, in 2023, the Centre had 9 PhD students, 12 research fellows, and 6 research assistants working in wet-lab and computational research. I also co-led the International Cancer Genome Consortium working group on mutational signatures. I am one of 2 corresponding authors on the resulting reference paper for these signatures, which has been cited > 3,200 times since 2020 https://doi.org/10.1038/s41586-020-1943-3).
I have wide-ranging experience in software development and data science. I developed and maintained the widely used Primer3 program for PCR primer design, and my lab has released several R packages on CRAN and GitHub.
Recipient of the 2018 American Association for Cancer Research Team Science Award. Used mutational signatures to discover widespread exposure to the carcinogen aristolochic acid in liver cancer (cited 380 times). Elucidated genomic landscapes of gastric cancer (cited 724 times), bile-duct cancer (3 papers, cited 1,490 times), and several other cancer types.
 |
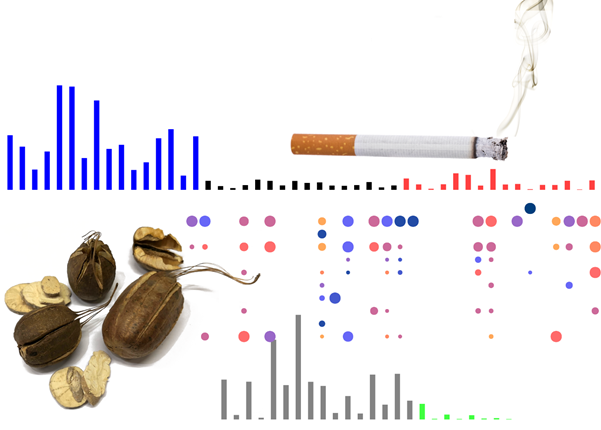
| Mutational signatures of cigarette smoking (blue, black, and red bars, top) and aristolochic acid (AA), a carcinogen in some herbal medicine (example dried herbs bottom left and the signature represented by grey bars, bottom). The circles in the middle provide a partial view of which cancer types have which signatures – each column is a cancer type; each row is a signature. |
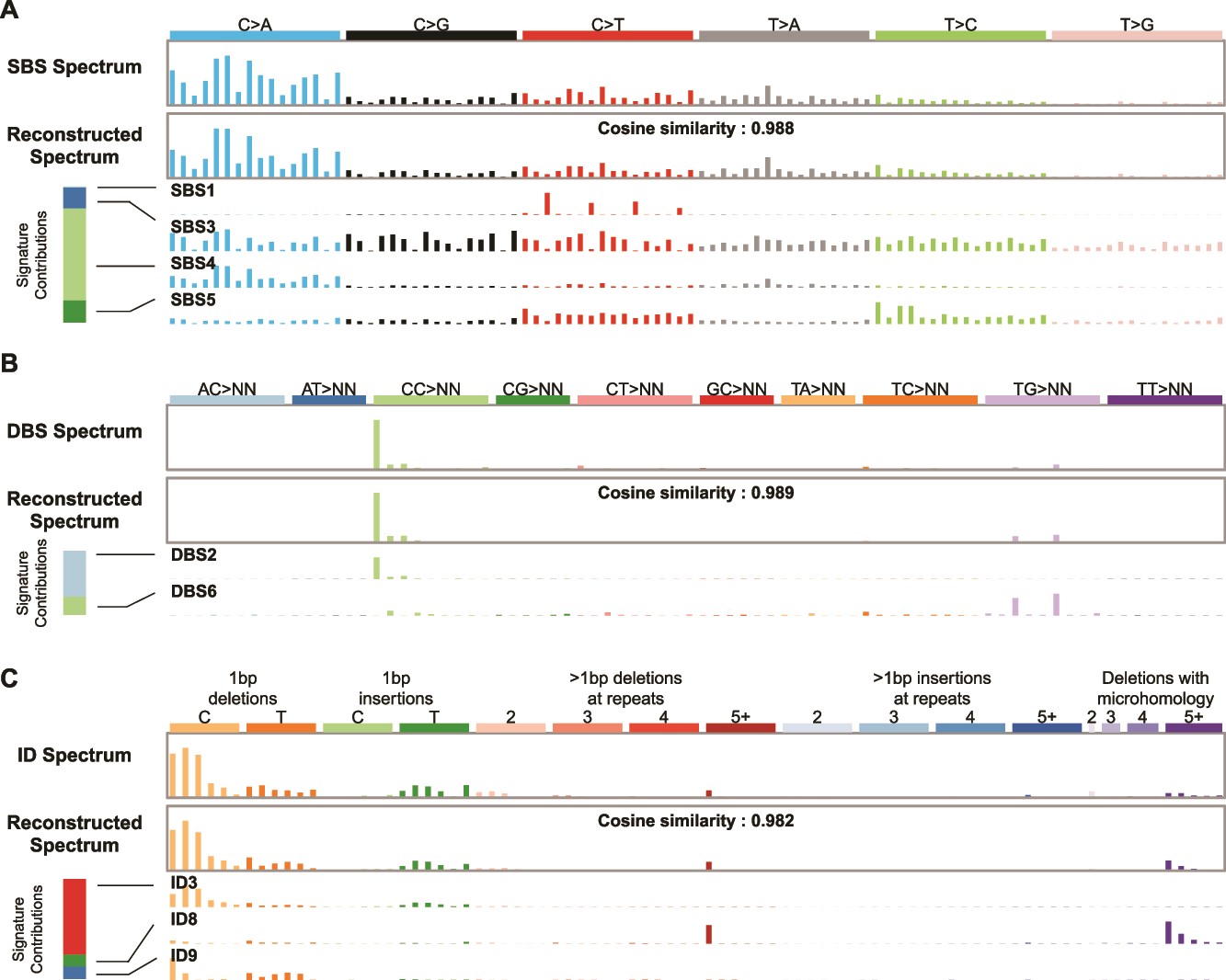 |
| Mutational signature attribution The task of mutational signature attribution is to find signatures that can reconstruct the mutational spectrum well. (A) Example of an SBS spectrum that can be reconstructed with a cosine similarity of 0.988 from four signatures. The bar at the left shows that mutational signature SBS4 contributed the most mutations to this spectrum. This signature is associated with tobacco smoking. (B) The DBS spectrum from the same tumor can be reconstructed with a cosine similarity of 0.989 from two signatures. The bar at the left shows that mutational signature DBS2, which is also associated with tobacco smoking, contributed the most mutations to this spectrum. (C) The ID spectrum from the same tumor can be reconstructed with a cosine similarity of 0.982 from three signatures. The bar at the left shows that mutational signature ID3 contributed the most mutations to this spectrum. Like the SBS signature SBS4, this ID signature is also associated with tobacco smoking. From "Benchmarking 13 tools for mutational signature attribution, including a new and improved algorithm" |
updated May 2025